Short-amplikonové sekvenování
Sequencing of PCR amplicons to study genetic variation within small target regions. The method involves two PCR steps:
- Amplification of specific locus using target-specific primers equipped with universal overhangs.
Performed by the facility user. - Reconstitution of sequencing adapters (artificial sequences carrying motifs necessary for the analysis) and incorporation of unique sequence
identifiers (barcodes / indexes).
Performed by NGSf staff.
General workflow
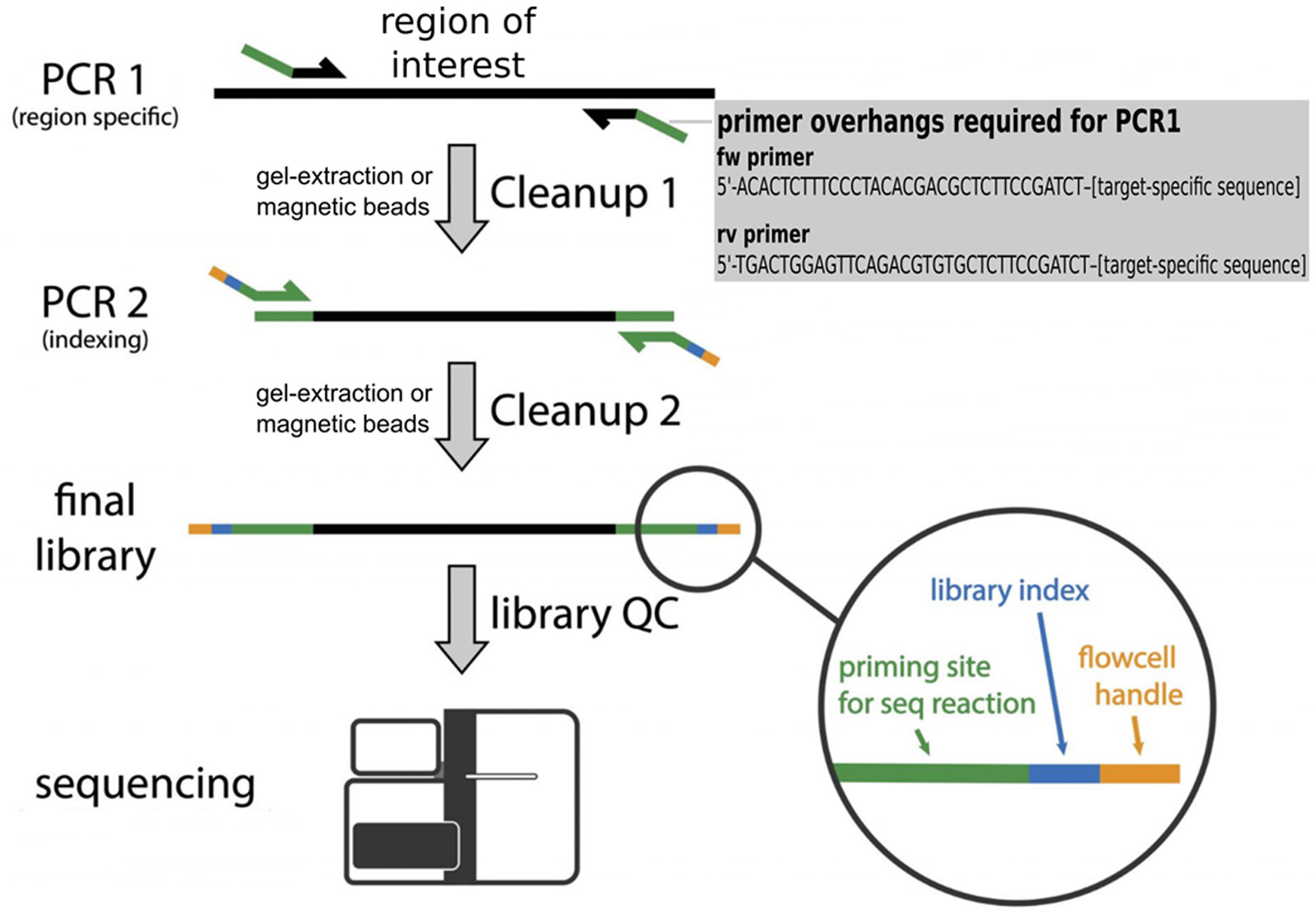
Overhang sequences for targert specific primers
FW primer: 5'-ACACTCTTTCCCTACACGACGCTCTTCCGATCT–[target-specific sequence (optimaly 20bp)]RV primer: 5'-TGACTGGAGTTCAGACGTGTGCTCTTCCGATCT–[target-specific sequence (optimaly 20bp)]
Sequencing analysis conditions
- Sequencing output: 2 × 150bp or 2 × 250bp Illumina paired-en reads (fragment sequenced from both sides)
- Data amount: 105 - 107 sequenced fragments (depending on requirements)
- Turnaround time: 2 - 3 weeks for 2 × 150bp, 3-4 weeks for 2 × 250bp
- Frequency: monthly
- Collection of samples: within first week of a month, shipment in the first part of following week
Sample submission guidelines
- Sample Type: Purified PCR fragment of interest (amplified with suitable primers), and eluted in water or TE.
- Sample Amount: >0.25 ng/µl.
- Sample Volume: 10 µl.
- Sample QC: Information about the concentration an volume + gel image or output from fragment analyzer.
- Maximum number of samples/plate: 94. When using a plate, leave the last two wells in column 12 empty (do not add water or buffer).
- Sample naming: samples must be described as follows “GRP_USR_01”, where GRP represents first 3 letters of the groupleader name, USR first 3 letters of the user name and last field is reserved for a 2 digits sample ID.
- Sample submission form: Please first register your project in request form, then fill your samples into sample form
Guidelines for PCR1 (performed by the user)
| Reaction | Cycling conditions | ||
| 5X Q5 Reaction Buffer | 5 µl | 98°C / 30 s | |
| 10 mM dNTPs | 0.5 µl | 98°C / 10 s | |
| 10 µM Forward Primer | 1.25 µl | 50–72°C⁑ / 10–30 s (⁑25 cycles) | |
| 10 µM Reverse Primer | 1.25 µl | 72°C / 20–30 s | |
| Q5 High-Fidelity DNA Polymerase | 1.25 µl | 72°C / 2 min | |
| Template DNA | variable* | 4–10°C / hold | |
| Nuclease-Free Water | to 25 µl | ||